Epigenetics field has undergone a “boom” in recent years due to new cutting edge technologies. It is focused on changes produced in the DNA and histones. How? Modifying chromatin structure whilst preventing any modifications from happening at the nucleotide sequence level, but producing changes at the level of gene expression. There are several modifications occurring to histones: methylation, acetylation, ADP-ribosilation, ubiquitination and phosphorylation.
- DNA methylation
- Chromatin immunoprecipitation (ChIP)
- Chromatrap isolates chromatin easily and efficiently
- How does it perform in the laboratory?
DNA Methylation has a crucial role in silencing gene expression and affecting all cellular processes. Changes in DNA methylation patterns are common during the first stages of cancer development or neurodegenerative diseases
Several methods have been developed to detect DNA methylation: bisulfite genomic sequencing, enzymatic digest, and chromatin immunoprecipitation (ChIP). Here, we are going to focus on chromatin immunoprecipitation (ChIP).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) studies the proteins binding to specific regions of genomic DNA. Thanks to this technique we can identify DNA-protein binding interactions in the cell nucleus. These interactions play a fundamental role in gene expression regulation.
Some of the major weaknesses of this technique are: the need to obtain large sample volumes due to sample leaking during washes; low signal combined with abundant noise and reproducibility complications.
More and more studies show how the binding of these proteins – either transcription factors or modified histones – occurs at specific DNA locations in cells, producing different methylation patterns depending on the studied disease.
Briefly, during ChIP assays, chromatin fragments (DNA-protein complex) are crosslinked to stabilized specific DNA-protein interactions. After that, chromatin is isolated and fragmented in sizes ranging from 100-500 base pairs, through sonication or enzymatic digestion. The resulting fragments are selectively immunoprecipitated using antibodies against the protein of interest. This is followed by the final separation of DNA and protein components.
Chromatrap isolates chromatin easily and efficiently
How? Chromatrap technology uses columns and microplates with a porous polymer filter at the base which bounds to Protein A or G. This proprietary format is unique.
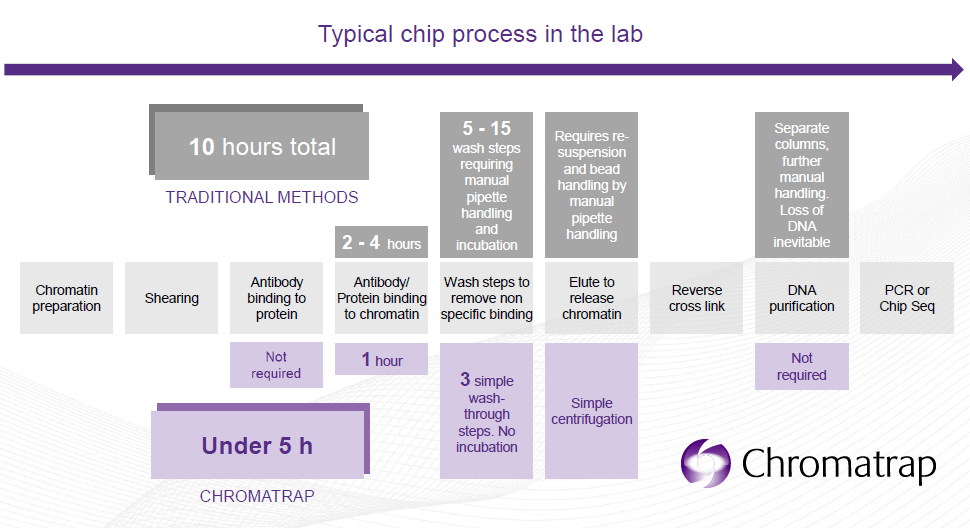
The novel solid state Chromatrap® protocols do differ from those used by other kit suppliers, including Active Motif, Diagenode, Cell Signalling Technology (CST), Abcam, Zymo Research and Epigentek, as these are all bead-based kits. During ChIP assays, the chromatin/antibody complex is selectively captured by the disks. 3 washes and an elution step are necessary to selectively enrich the DNA-protein complex of interest.
This process developed by Chromatrap is more efficient and advantageous than other techniques:
- ChIP in less than 5 hours: from sample preparation to qPCR.
- Less sample handling steps are required à preventing sample loss due to more pippetting steps.
- Single format assays and 96 well plates available.
- Optimized for smaller samples, providing better results in ChIP-qPCR.
- More assays are allowed per sample.
How does it perform in the laboratory?
Chromatrap® outperforms other chromatin immunoprecipitation assays because it has:
- Robust signal to noise- typically 2-3 times better than standard methods
- Low non-specific binding on the inert Chromatrap® discs
- No DNA clean-up required for qPCR
- High surface area and molecular mixing ideal for low abundant targets and challenging cell types
- Wide dynamic range makes it suitable for ChIP-seq
- 96 well format for high throughput analysis
- Allows more ChIP assays to be performed from a single sample
For more information, have a look at the following video.
Further information about Chromatrap, here.
Register to our newsletter here and we will keep you posted





Leave a reply