Whether you perform qPCR for the first time or just want to be sure you maximize your chances for success, there are a few basic principles to be aware of. Here, we will describe the top 5 rookie qPCR mistakes and how to best avoid them:
- 1. Poor quality RNA.
- 2. Selecting the detection chemistry
- 3. Not using a master mix and organizing your experiments.
- 4. Skimping on controls.
- 5. Not validating your reference gene
1. Poor quality RNA.
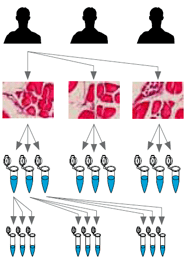 This is arguably the most critical step for the successful preparation of your cDNA and being able to perform efficient and reproducible RT-qPCR. RNA is extremely sensitive to degradation by RNases and must be handled with care during nucleic acid isolation steps.
This is arguably the most critical step for the successful preparation of your cDNA and being able to perform efficient and reproducible RT-qPCR. RNA is extremely sensitive to degradation by RNases and must be handled with care during nucleic acid isolation steps.
Degraded or contaminated RNA can negatively affect the efficiency and yield of your RT-qPCR experiment. Your lab bench, pipettes and tips must be RNase-free, and extracted RNA must be stored in an RNase-free solution.
When you assess RNA purity using the spectrophotometer, the ratio of the absorbance at 260 and 280 nm (A260/280) should be in the range of 1.8-2.0. If lower, this may suggest contamination of your sample with phenol or proteins.
2. Selecting the detection chemistry
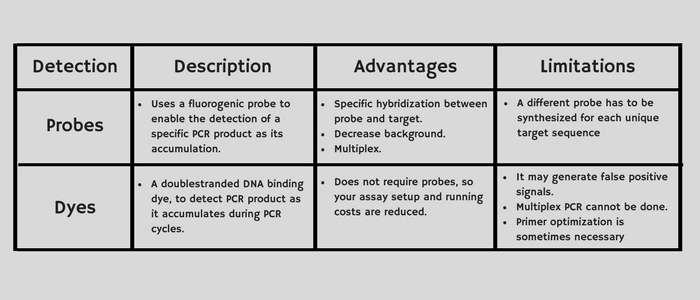
Every real-time PCR contains a fluorescent reporter molecule—a TaqMan® probe or SYBR® Green dye, for example—to monitor the accumulation of PCR product. As the quantity of target amplicon increases, so does the amount of fluorescence emitted from the fluorophore.
he two types of chemistries that have been developed for gene expression studies using real-time PCR are:
- TaqMan chemistry (also known as “fluorogenic 5´ nuclease chemistry”)
- SYBR Green I dye chemistry.
TaqMan probe-based chemistry and SYBR Green I dye can be used for the assay types listed below.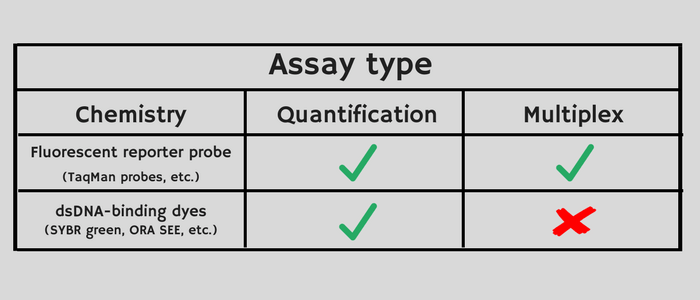 Each chemistry has its advantages and limitations. For example, TaqMan chemistry enables you to perform multiplex PCR. If high sensitivity is your priority, SYBR Green chemistry offers that advantage. Consider the following aspects of each chemistry type when choosing between TaqMan probe-based and SYBR Green chemistry for your assays:
Each chemistry has its advantages and limitations. For example, TaqMan chemistry enables you to perform multiplex PCR. If high sensitivity is your priority, SYBR Green chemistry offers that advantage. Consider the following aspects of each chemistry type when choosing between TaqMan probe-based and SYBR Green chemistry for your assays:
3. Not using a master mix and organizing your experiments.

Using a master mix for your reagents minimizes experimental variability and thus improves reproducibility by reducing well-to-well and sample-to-sample variations. Similarly, it is critical to stay organized in the design and execution of your qPCR experiments. This means making a table of your primers and cDNA to know what is going where. Nothing’s worse than getting distracted and for4etting where you have and haven’t pipetted, so it is essential to have an organized system in place for your pipetting. Some researchers choose their design in a pattern, such as setting up primers in alphabetical order.
This way, as you load your cDNA, you can match the tips to the wells a la Battleship-style (i.e., tip C7 to well C7 and so forth). No matter the pipetting strategy, don’t forget to spin your plate down at the end to get any liquid off the well walls.
4. Skimping on controls
It is important to include the appropriate controls in setting up your RT-qPCR assays. You should include a negative control that lacks your template RNA or cDNA and replace the volume with nuclease-free water, termed the “no template control” or NTC. This can detect cross contamination of surfaces or reagents (i.e. master mix, primers) as well as primer dimer formation. Thus, if you see amplification in this sample, you should run a gel to check for primer dimer or your target. In addition, you should run a RT negative control, where there is no reverse transcriptase, termed a “No amplification control” or NAC. If product is observed, this suggests DNA contamination of your sample.
5. Not validating your reference gene

The amplification of an endogenous control (or reference) gene allows for normalization of target gene expression by comparison of CT values. This can account for differences in the amount or quality of starting material (RNA or cDNA) as well as differences in RNA preparation methods or cDNA synthesis. A reliable reference gene is one whose expression level is unaffected by your experimental variables or interventions and does not differ between the relevant physiological states of your sample conditions.
¿Tienes dudas?
Si no te queda claro del todo cómo funciona esta tecnología, o quieres que te ayudemos a configurar tu ensayo, nuestro departamento técnico de especialistas, con amplia trayectoria en investigación (todos PhD), te pueden echar una mano: por mail (tecnic@labclinics.com), por tlf +34.934464700 o de forma presencial. Contáctanos y estaremos encantados de poder ayudarte!




Leave a reply