Epigenetics studies changes in gene expression that do not involve alterations in the DNA sequence. The prefix epi- means “above” or “in addition to”, therefore, the word epigenetics encompasses all those processes above genetics and that modify gene expression.
DNA methylation

The enzymatic addition of methyl (CH3-) groups to the DNA molecule is called DNA methylation. In mammals, methyl groups are added almost exclusively to cytokines in CpG dinucleotides. There are genomic regions with a high content of CpGs and are called CpG islands.
Abnormal changes in DNA methylation have been identified in many different diseases such as cancer, inflammatory, autoimmune, psychiatric, cardiovascular, and age-related diseases. When these CpG islands are found in the promoter regions of the gene, DNA methylation acts to repress gene transcription.

Sodium bisulfite treatment
During DNA replication, the enzyme DNA Methyl-Transferase (DNMT) re-synthesizes the methyl groups on the newly replicated strand. In such a way that if we carry out a PCR, these enzymes will not be present in the reaction and therefore the amplified locus will lose the information regarding the methylation state. In order to analyze the methylation pattern of cytosines within the locus of interest, information about which cytosines are methylated must be preserved before performing PCR amplification. Sodium bisulfite treatment deaminates unmethylated cytosines into uracils while leaving methylated cytosines intact (in other words, methylated cytosines are resistant to sodium bisulfite-induced modifications).
The melting temperature of DNA
The dissociation of the DNA double helix is called DNA fusion. The melting temperature of DNA depends on the number of hydrogen bonds that the complementary strands contain. Between adenine (A) and thymine (T) there are two hydrogen bonds while between guanine (G) and cytosine (C) three bonds are formed. Therefore, the more G and C present in the sequence, the higher temperature will be needed to melt a piece of DNA.
How to detect differences in melting temperature
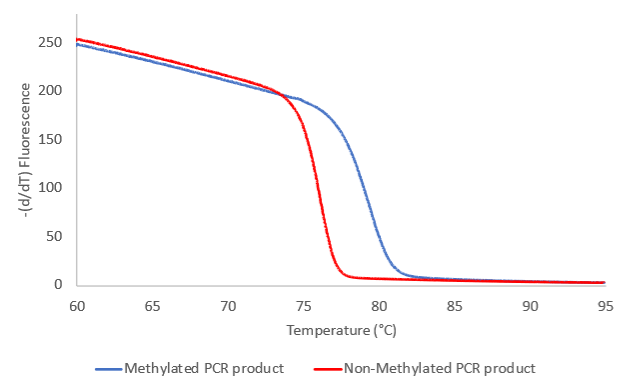
The most commonly used method to determine the melting temperature of a PCR product is to subject it to a temperature gradient in the presence of an intercalating dye. These intercalating agents emit light upon binding to double-stranded DNA.

On the left the result of the emitted fluorescence changes is shown. At the beginning of the experiment, at low temperatures, the fluorescence is high and therefore the majority of the PCR product is double-stranded (A). As we increase the temperature, the emitted fluorescence slowly decreases, until a temperature in which the hydrogen bonds are broken drastically. (B). At high temperature there are no double stranded PCR products in the sample and fluorescence levels are close to 0 (C).
The temperature at which we observe the sharp drop in fluorescence depends on the number of hydrogen bands in the analyzed PCR product and is therefore specific to the analyzed fragment.
How to analyze the results by High Resolution Melting
In principle, after PCR amplification, a C-rich PCR product is obtained if the amplified locus was methylated (see graph to the right) and a T-rich PCR product if the locus was unmethylated. These PCR products will show different melting profiles. The amplified PCR product from the unmethylated version of the specific locus will have a relatively low melting temperature and will melt earlier down the temperature gradient than the amplified PCR product from the methylated version of the same locus.
Presence of controls in methylation detection experiments
Following good laboratory practices, any experiment must include its respective controls. In PCR-based methylation detection experiments, two types of controls are needed: the methylated (positive control) and the unmethylated (negative control) sample. These controls are typically chemically modified DNA samples consisting of methylated and unmethylated versions of the locus of interest and are reference points for analysis of an unknown sample.

In principle, hypomethylation is observed if the melting profile of a sample does not overlap with the methylation profile of the positive control, and hypermethylation is observed when the melting profile of the sample does not overlap with the melting profile of the unmethylated control. .
Methyldetect offers tests to see whether or not there is methylation for a wide catalog of genes. Also, if your study object is a different gene than those on the list, we can custom design the kit.
Both to request a different gene and to request more information or a quote, you can do so in the following form:




Leave a reply